|
Deep Learning Enables Cellular Behavioral Prediction and Label-Free Detection by Cell Morphology
Cell morphological analysis is one of the oldest methods in cell biology, yet people conventionally focus on few morphological features with limited images. Using our high-throughput single-cell migration chip and deep learning models, we initiated research to correlate morphological features of cellular contours and organelles (e.g., nucleus and mitochondria) with cell migration. In addition, we developed a fast, low-cost, and label-free method to assess tumor sphere viability for large-scale drug screening in microfluidics. Recently, we also developed a Live, Apoptotic, and Necrotic Cell Explorer (LANCE) to categorize cell death status in a label-free manner. The non-destructive label-free LANCE method allows for live tracking time dynamics of the cell death process. More interestingly, we developed a future-telling model to estimate the Day 14 experiment outcomes based on Day 4 images. The method significantly expedites tumorsphere assays for the investigation of cancer stem-like cells (CSCs). The combination of microfluidic single-cell analysis and machine learning provides a high-throughput, fast, low-cost, and non-destructive analytical method to enhance bio-discovery. “LANCE: a Label-Free Live Apoptotic and Necrotic Cell Explorer Using Convolutional Neural Network Image Analysis,” Analytical Chemistry, 94(43), 14827-14834, 2022 (cover). [Link] “Early Prediction of Single-cell Derived Sphere Formation Using Convolutional Neural Network Image Analysis,” Analytical Chemistry, 92, 11, 7717–7724, 2020. [Link] “Morphological Prediction and Feature Identification of Cell Migration Using Artificial Neural Network and Random Decision Forest,” Integrative Biology, 10(12), 758-767, 2018. [Link] "Label-free Estimation of Therapeutic Efficacy on 3D Cancer Spheres Using Convolutional Neural Network Image Analysis," Analytical Chemistry, 91, 21, 14093-14100, 2019. [Link] |
|
Migration-Based Cell Sorting for Discovering Novel Cancer Regulators
We developed high-throughput microfluidic cell migration platforms that coordinates robotic liquid handling and computer vision for rapidly quantifying the movement of hundreds of thousands of individual cells. In addition to phenotypic observation, we performed migration-based cell sorting using a microfluidic migration chip. Given the hypothesis that only some cancer sub-populations are likely to metastasize, we aim to find the metastatic ones and figure out their molecular features as therapeutic targets. While conventional cancer biology distinguishes cellular heterogeneity by bio-marker systems, these systems suffer from inconsistency between patients and malignancies. More importantly, standard markers do not directly relate to actual malignant cell behaviors (e.g. invasion). Using the microfluidic cell migration chip we developed, we separated cells based on migration speed. Notably, we found a small sub-population of highly migratory cells having significantly greater tumor initiation and metastasis capability in a mouse xenograft model. We further analyzed this migratory sub-population using whole transcriptome sequencing and uncovered a gene called phosphatidylserine decarboxylase (PISD), which regulates tumor-initiating cell through altering mitochondrial morphology and functions. Our research establishes not only the genes associated with cancer metastasis, but also an innovative approach to identify novel cancer regulators using a microfluidic cellular behavioral assay. “Microfluidic Single-Cell Migration Chip Reveals Insights into the Impact of Extracellular Matrices on Cell Movement,” Lab on a Chip, 23(21), 4619-4635, 2023. (cover). [Link] “High-throughput cellular heterogeneity analysis in cell migration at the single-cell level,” Small, 2206754, 2022. (cover) [Link] “Single-cell RNA-sequencing of migratory breast cancer cells: discovering drivers of cancer metastasis,” Analyst, 144, 7296 – 7309, 2019. [Link] “Functional Isolation of Tumor-Initiating Cells through Microfluidic-Based Cell Migration,” Scientific Reports, 8, 244, 2018. [Link] “Single-Cell Migration Chip for Motility-based Microfluidic Selection of Heterogeneous Cell Populations,” Scientific Reports, 5, 9980, 2015. [Link] |
|
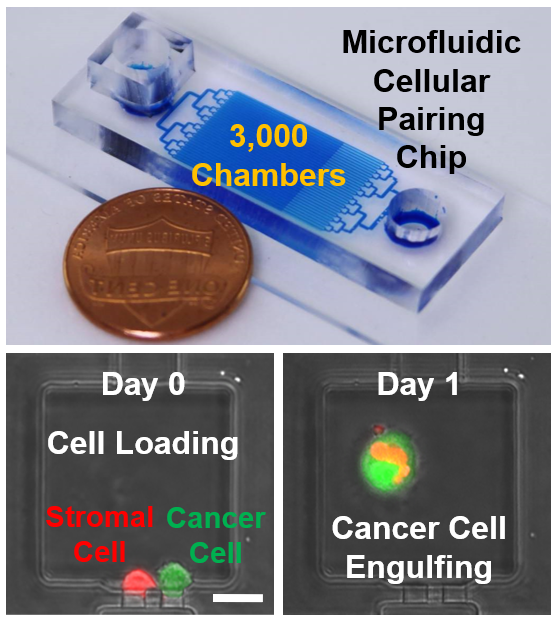
Microfluidic Cellular Pairing and Retrieval for Studying Breast Cancer-Stromal Engulfment
Malignant cells have been the focus of cancer biology, yet this approach overlooks effects of stromal cells on tumor progression and response to therapy. Mesenchymal stem/stromal cells (MSC) in a tumor microenvironment stimulate breast cancer metastasis, but the exact mechanisms are still controversial. Using an innovative microfluidic platform, we paired single cancer cells with single stromal cells in micro-chambers in high throughput. With a large number of cell pairs, for the first time, we directly observed the rare process of cancer engulfment of stromal cells. In addition to qualitative observation, we quantified cellular engulfment rates and found that metastatic breast cancer cells have elevated capacity to engulf MSCs as compared to non-metastatic breast cancer or non-tumorigenic cells. Furthermore, we used a novel photo-acoustic cell retrieval technology to selectively retrieve engulfing cancer cells and compare them with wild-type ones. We found the engulfment of MSC promotes EMT and acquisition of stem cell traits, which potentiates the migratory, invasive, and metastatic ability of breast cancer cells. Whole transcriptome analysis uncovered a signature of previously unconsidered genes and pathways associated with MSC engulfment and metastatic dissemination. These results pinpoint a new mechanism by which MSCs in the tumor microenvironment promote breast cancer metastasis and a new microfluidic test with potential utility in the clinic to identify metastatic ability of breast cancer cells. “Mesenchymal stem/stromal cell engulfment reveals metastatic advantage in breast cancer,” Cell Reports, 27, 3916–3926, 2019. [Link] “A hybrid breast cancer/mesenchymal stem cell population enhances chemoresistance and metastasis,” JCI Insight, e164216, 2023. [Link] |
|
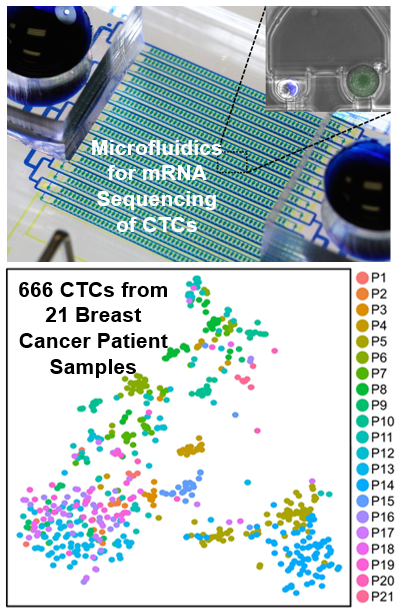
Single-Cell Sequencing of CTCs for Precision Medicine
Analyzing individual circulating tumor cells (CTCs) as a liquid biopsy has high potential for diagnosis, assessment of prognosis, and treatment in cancer, yet blood samples contain merely tens of cancer cells mixed with millions of WBCs and billions of RBCs. Though investigators have developed various methods to enrich CTCs in blood sample, the challenges of analyzing small number of cells amidst large numbers of residual contaminating cells are still far beyond capabilities of existing tools. To address the challenges, we developed a hydrodynamic microfluidic platform (HydroSeq) that removes contaminating blood cells while isolating ~70% of CTCs for single-cell RNA sequencing. We have processed 21 blood samples from breast cancer patients and successfully sequenced 666 CTCs, which are one order of magnitude more than previous studies. Thanks to the large amount of data we acquired, we uncovered the heterogeneous nature of CTCs. Remarkably, different CTC populations (e.g. HER2- epithelial-to-mesenchymal transition (EMT)-like and HER2+ MET-like) can co-exist within a single patient sample. The observed heterogeneity suggests specific targets for therapy that would not be captured by sequencing many CTCs in bulk or with WBC/RBC contamination. “Hydro-Seq Enables Contamination-free High-throughput Single-cell RNA-sequencing for Circulating Tumor Cells,” Nature Communications, 10, 2163, 2019. [Link] |
|
|
|
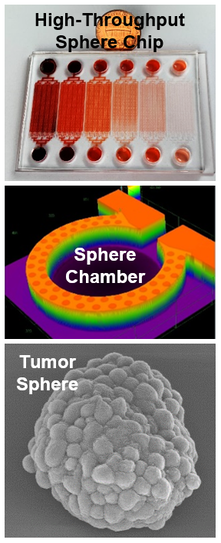
High-Throughput 3D Culture of Cancer Spheres
Despite recent advances in cancer treatment, developing better therapeutic reagents remains an essential task for oncologists. To accurately characterize drug efficacy, 3D cell culture holds great promise as opposed to conventional 2D monolayer culture. Using microfluidic technology, we have developed various 3D cell culture platforms for high-throughput screening of chemotherapeutic drugs. In addition, single-cell 3D culture is valuable for tumorsphere assay. Tumorsphere assay is an established method to identify cancer stem-like cells (CSCs) functionally and study this sub-population critical for cancer progression and metastasis. The method is based on the unique capability of single CSCs to survive and grow to tumorspheres in harsh suspension culture environment. However, conventional method of tumorsphere assay in low-attachment plate/dish suffers from high variation, low reproducibility, and low throughput. To address the challenges, we used microfluidic technology to isolate thousands of single cells in suspension for high-throughput tumorsphere assay. “Scalable Multiplexed Drug-Combination Screening Platforms Using 3D Microtumor Model for Precision Medicine,” Small, 14, 42, 2018. [Link] “Scaling and Automation of High-Throughput Single-Cell- Derived Tumor Sphere Assay Chip,” Lab Chip, 16, 3708 - 3717, 2016. (cover) [Link] “High-Throughput Single-Cell Derived Sphere Formation for Cancer Stem-Like Cell Identification and Analysis,” Scientific Reports, 6, 27301, 2016. [Link] “High-Throughput Cancer Cell Sphere Formation for Characterizing the Efficacy of Photo Dynamic Therapy for 3D Cell Cultures,” Scientific Reports, 5, 12175, 2015. [Link] |